SMAP-Selective Molecular Assembly Patterning
Project
The project goal is the development of a new chemical patterning technique in the nanometer range for biological applications. Selective Molecular Assembly Patterning is based on photolithographic processes to create patterned SiOn2/TiO2 substrates with subsequent selective adsorbtion of protein- and cell-adhesive and -resistant organic molecules. Since it is not the organic molecules that are patterned, but rather the metal oxide substrates, SMAP can yield contaminant-free, nanometer-scale-patterned, large-area surfaces.
Materials and Methods
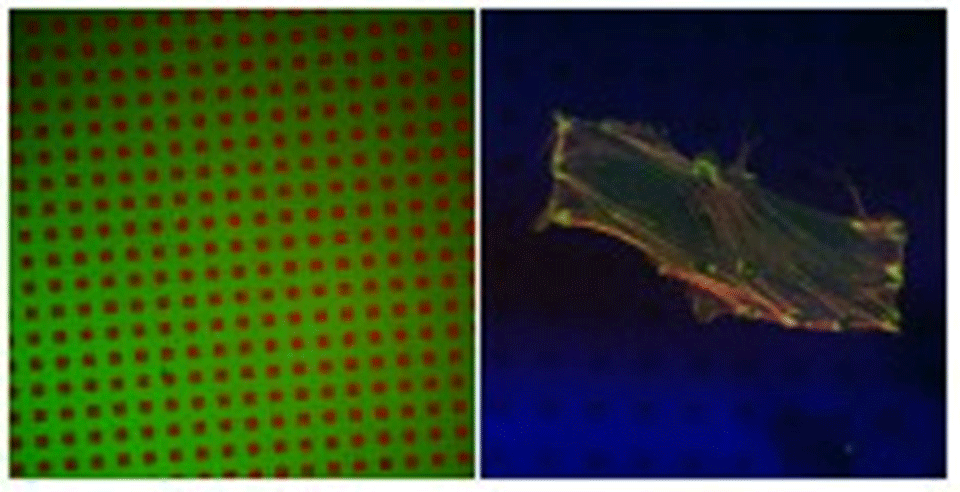
Standard photolithography was used to create patterns of titanium oxide within a matrix of silicon oxide. Using the fact that ordered SAMs of alkane phosphates form on the TiO2, but not on the SiO2, surfaces, by self-assembly from aqueous solutions, the TiO2 structures were rendered hydrophobic and hence protein-adsorbing. Poly-L-lysine-g-poly(ethylene glycol) (PLL-g-PEG)3,4 was used to render the exposed SiO2 protein-resistant, thus creating a contrast with respect to protein adsorption. X-ray photoelectron spectroscopy and imaging time-of-flight secondary ion mass spectrometry were used to characterize the surfaces in vacuo, while fluorescence microscopy (Figure 1A) was used for studies in aqueous media.
Results
Quantitative XPS as well as qualitative ToF-SIMS results proved that dodecylphosphate (DDP) adsorbed on the TiO2 surface, forming a self-assembled monolayer5 and leaving SiO2 bare. Subsequent modification with PLL-g-PEG resulted in protein-adhesive patches (TiO2/DDP) within a matrix that is resistant to protein adsorption (SiO2/PLL-g-PEG), as shown in Figure 1A. Human foreskin fibroblasts (HFF) as well as smooth muscle cells (SMC), incubated with such substrates in serum, exhibited a clear preference towards attaching to the protein-adhesive (hydrophobic) areas (Figure 1B), where they form focal contacts.
References
1Wheeler et al.(1999) J Biomech Eng 121:73-78. 2Chen et al. (1997) Science 276: 1425-1428. 3Elbert and Hubbell (1998), Chem Biol 5:177-83. 4Kenausis et al. (2000) J Phys Chem B 104:3298-3309. 5Textor et al. (2000) Langmuir 16:3257-3271.
Partners
- Institute for Biomedical Engineering, IBE, ETH Zürich
- external page Dr. h. c. Robert Mathys Foundation, CH-2544 Bettlach
- external page Institut Straumann AG, CH-4437 Waldenburg
- external page Zeptosens AG, CH-4108 Witterswil
Funding
- TOP NANO 21 Program of the ETH-Council, KTI Nr. 4597.1